1 前言
源自于历史时期生物残留的古DNA,为重塑不同地理区域古生物变迁、人类进化等提供了带有时间刻度的遗传信息[1-2],特别是高通量测序技术、DNA捕获技术和宏基因组分析的出现,极大提升了古DNA分析的准确性和研究范围[3]。相较于依赖古生物化石等载体只能获得单一物种的古DNA,从历史时期埋藏的沉积物中提取的古DNA不仅可提供动物、植物、真核生物、原生生物和原核生物等极大生物分类学宽度的动态信息,还可提供相关生物生存的地理环境信息[4⇓-6]。通过在永冻、干燥、厌氧、淋溶作用弱的环北极冻土区[7-8]、欧洲高山湖泊流域[9]、稀有的干燥洞穴[10]等地理环境中开展沉积物古DNA研究,相关学者将古DNA指标广泛用于古气候和古环境重建[7,11⇓ -13]、生态系统及功能服务[14⇓-16]、物种变迁及环境效应[8,17 -18]、农牧活动及人地关系演变[9,19]、环境考古及人类饮食[10,20⇓ -22]等研究中。例如,Kjær等[23]基于北格陵兰岛Kap København地层中的沉积物古DNA组成,发现了大块化石和孢粉组合未记录的生物遗存,重现了已消失的200万年前更温暖气候环境中一个开放的北方森林生态系统的演变。然而,国内的沉积物古DNA研究极为有限,中文文献更多是成果综述和展望[22,24⇓⇓⇓⇓ -29],但也有与国外机构合作开展了植被更替、生态环境演变的部分研究[7,30⇓ -32],如高玉等通过与丹麦哥本哈根大学地质遗传中心合作开展的青藏高原环境考古的研究,基于考古遗址沉积物古DNA组成发现了籼稻在青藏高原南部的传播路径[33],并揭示了廓雄遗址的食物构成[34]。近期,Zhang等[35]基于太湖沉积物DNA组成重建了蓝藻群落近百年变化过程,指出微孢藻属丰度的变化主要受控于温度和风速等自然环境要素,进而削弱全球变暖背景下人们对湖泊富营养状况的改善效果。这均表明国内沉积物古DNA研究已逐渐起步[30,35]。
虽然沉积物古DNA指标可为理解中国不同气候环境下多样地理空间的生态环境演变及人地关系提供基因视角[5,28],但国际上对古DNA的数据解释仍存在诸多争议[4]。例如,Giguet-Covex等[36]分析了法国阿尔卑斯山北部Anterne湖泊现代沉积物DNA组成,虽该湖泊流域目前以羊群放牧为主,但在现代沉积物DNA中并未识别出羊的核外DNA。上述结果的出现源自对流域范围内沉积物DNA的地表过程未有清晰的认识[4],限制了对DNA沉积过程的深入理解。目前,国际上对于沉积物DNA在流域范围内的地表过程仅形成以下两方面的初步共识。① 生物来源:湖内沉积物DNA部分来自于湖内生物的沉淀,陆源生物的输入主要集中于滨湖区、河流下游等。因此,沉积物DNA指代的陆源生物类别具有“本土”属性[5]。② 输移和保存过程:土壤理化属性(如黏土和有机质含量、矿物组成)和河流水文特征(pH、氧化还原、河流连通度、电导率、离子浓度等)均影响流域表土DNA向湖内输移的比例和损伤状况[36]。例如,Jia等[32]研究表明100~500 μS/cm电导率和7~9 pH值的湖水环境是研究湖泊沉积物DNA的适宜环境。因此,在将沉积物DNA这一分子生物学指标应用到重建中国多样的第四纪生态环境之前,亟需开展不同气候和沉积环境下表土DNA埋藏过程及影响因素的对比分析,以期实现对不同时空背景下沉积物DNA组成的科学解释[37-38]。
沉积物DNA浓度鉴定是基于DNA组成开展生物信息分析流程的首要环节,决定着后续实验开展的必要性[25,29]。而地处“地球第三极”青藏高原东北部青海湖盆地的布哈河流域,海拔高达3000 m以上,气候寒冷干燥,且DNA来源丰富、土地利用方式多样,是研究表土DNA地表过程的理想地理位置。因此,本文基于国内搭建的沉积物DNA实验平台,以布哈河流域表土为载体,分析该流域内表土DNA浓度的空间分布状况,并结合沉积物的理化组成、流域气候、植被和土地利用信息,揭示该流域表土DNA浓度的受控因素,为理解布哈河流域沉积物DNA的埋藏过程及其环境效应提供科学依据,也为国内沉积物古DNA指标在第四纪环境研究的广泛应用提供参考。
2 研究区概况
布哈河流域(36°15′N~38°20′N, 97°50′E~101°20′E)位于青藏高原东北端的青海湖盆地西北部,北临大通山,南临青海南山,东临日月山,西临阿木尼尼库山,流域面积为1.43×104 km2,海拔高度范围为3036~5068 m(图1)。布哈河发源于疏勒南山,流经天峻县城和共和县吉尔孟乡等地,沿西北—东南方向汇入青海湖。根据布哈河流域地形地貌和海拔梯度变化,源头至快尔玛乡河段为布哈河上游,快尔玛乡至吉尔孟乡河段为中游,吉尔孟乡至汇入湖泊口的河段为下游[39]。作为青海湖最大的入湖河流,布哈河干流长度286 km,主要支流有艾热盖河、希格尔曲、夏日格曲、江河、吉尔孟河等,年均径流量达9.08×108 m3,年均输沙量为3.61×105 t,是青海湖西侧主要泥沙来源。布哈河水pH为8.22,盐度为0.09~0.13,其间含有大量碳酸盐,属于高盐度淡水水质[40]。布哈河中主要鱼类为国家二级保护动物裸鲤,河水的淡水水质为裸鲤的季节性洄游提供了天然场所[41]。布哈河流域属高原半干旱季风气候区,多年平均气温为-8.5~1.6 ℃,年均降水量为268~510 mm,5—9月降水量占全年降水的61.84%(图1),无霜期仅有95 d[42]。流域内土壤类型多样,主要有高寒冻沙土、高寒草甸土、高寒草原土等[43],呈季节性冻土,冷冻时长约为7个月(当年10月下旬到次年4月),最冷时冻土深度达1 m左右[39]。流域内主要植被类型为高寒草甸、高寒草原、高寒流石坡稀疏植被等,优势种为蒿属植物。经实地调查,布哈河上游区域植被覆盖度较低,主要以草甸为主;中游区域河谷中常见低矮灌木丛(以沙棘丛为主),并在人类居住区伴有一些荒废的耕地;下游植被覆盖度最高,但更多被当地牧民整改为适用于放牧的高盖度草地。布哈河流域自古以来即为当地牧民的天然牧场,野生动物和家养畜牧动物种类丰富,主要有牦牛、绵羊、黄牛、普氏原羚、高原鼠兔等,放牧规模达70头/km2以上,是当地居民的主要经济来源[44]。
图1
图1
青海湖布哈河流域地理位置及采样点分布
Fig. 1
Geographic location and the distribution of topsoil samples in the Buha River catchment, Qinghai Lake
3 样品采集和实验方法
3.1 样品采集
基于对布哈河流域详细的野外调查,于2021年5—6月沿源头—入湖口方向在布哈河流域上游、中游、下游的干流河谷及主要支流江河、吉尔孟河等采集沿岸表土样品共22个,标记为单数号,样品采集间隔大体为15~20 km左右。具体而言,在布哈河上游采集样品6个、中游4个、下游4个、江河3个、吉尔孟河5个(图1)。样品采集后立即保存于4 ℃的恒温冷藏柜中,确保样品中的DNA后期无降解。因其中6个沿岸表土样品距离较近,故选取间隔较为一致的16个土样开展表土DNA浓度分析。
3.2 表土DNA浓度测定
在DNA提取超净实验室,先称取5~10 g土样装入50 mL离心管中,以1∶2的比例添加配备好的磷酸缓冲液(phosphate buffer),并摇匀15 min以分离土壤中吸附的DNA[46-47]。经过10 min离心后,移取4 mL上清液至超滤离心管中进一步浓缩土壤样品中的DNA。随后,利用德国Macherey-Nagel公司生产的NucleoSpin®土壤DNA提取试剂盒(Soil Kits),完成DNA浓缩液提取、粘合、洗涤、干燥、DNA洗脱的步骤[4,29,48]。在此过程中,加入空白控制样同步完成上述步骤,以明确实验过程中的污染状况。具体而言,经对超滤离心管10 min离心后,生成至少400 μL的DNA浓缩液;在添加了200 μL的SB buffer后,高速离心1 min将浓缩液中的DNA粘合到滤膜上;随后利用SW1和SW2 buffer对滤膜进行洗涤,并通过2 min的高速离心干燥滤膜;最后加入SE buffer将滤膜上的DNA洗脱到收集管中,且冷冻保存在-20 ℃的冷柜中,便于后期分析。在完成了土壤样品DNA提取的基础上,先根据凝胶电泳法利用LabChip GX Touch微流控毛细管电泳系统进行核酸完整度分析,随后基于紫外分光光度法利用NanoDrop分光光度计测定各样品的DNA浓度,测试误差小于4%[49]。最后,基于下列公式对测定的DNA浓度值转换为质量浓度。
式中:Csoil DNA是土壤DNA的质量浓度(μg/g);Ct为测定的浓缩液中DNA浓度(ng/μL);L是超滤过程中DNA浓缩液体积(μL);Msoil是离心管中的土壤质量(g)。
3.3 粒度、元素、烧失量和TOC分析
将所有表土样品冷冻干燥后,过20目筛除去可见植物残体和大颗粒杂质。称取0.4 g左右的过筛土样并添加浓度为10%的双氧水(H2O2)和稀盐酸(HCl)分别去除土壤中的有机质和碳酸盐,静置48 h后加入六偏磷酸钠((NaPO3)6)分散剂,利用英国马尔文公司生产的Mastersizer 2000激光粒度仪测定土壤样品的粒度组成,测试误差小于2%。随后,利用玛瑙研钵将5 g过筛土样研磨至200目。取0.6 g研磨土样熔融后利用荷兰帕纳科公司生产的Axios advanced(PW4400)X荧光光谱仪定量分析样品的元素组成,参数设置为30 ℃的检测环境、750 Kpa大气压及Fuse09001-2的分析方法,随后利用不同元素组分间的比例计算淋溶系数(β = Si2O/(MgO+CaO+K2O+Na2O))和化学蚀变指数(CIA = [Al2O3/(Al2O3+CaO+K2O+Na2O)]×100%)[50]。表土样品的烧失量(LOI)利用煅烧法在马弗炉中完成。称取1 g研磨样品放置在干燥的瓷舟中,在550 ℃和950 ℃恒定温度下分别煅烧4 h和2 h,利用高精度天平(精度为0.0001)称量和计算各温度下损失的土壤重量。最后,取1 g研磨土样添加10%稀盐酸去除样品中的无机碳后,使用德国Elemertar公司生产的vario TOC cube总有机碳分析仪测定土样的TOC含量,测试偏差低于10 %[51]。
3.4 空间数据处理
布哈河流域的DEM数据源自地理空间数据云(
4 实验结果
4.1 表土DNA浓度分布
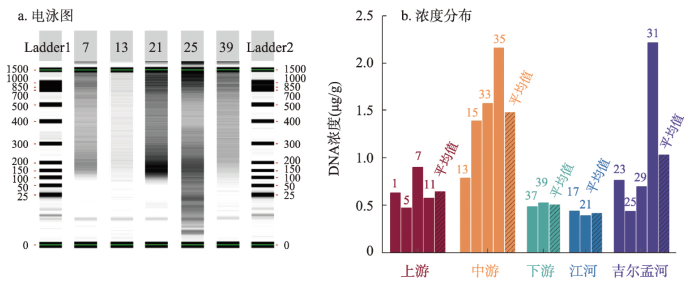
布哈河流域表土DNA浓度主要由两步骤确定,即核酸完整度分析和浓度测定。核酸完整度分析主要用于检视表土中DNA的保存状况,进而明确DNA的鉴别方法[25]。选取干流上、中、下游及支流江河、吉尔孟河段各一个土样,以其电泳图反映布哈河表土的核酸完整度和DNA保存状况。结果表明,该区域表土的核酸电泳图均存在明显拖尾现象,核酸组成存在较严重的降解(图2a),不能用来直接识别DNA指示的生物类型。另一方面,布哈河流域的表土DNA浓度范围为0.39~2.21 μg/g,平均值为0.91 μg/g,最高值和最低值分别出现在吉尔孟河和江河的汇入口(图2b),可能与两个点的土地利用有关。就河段分布而言,干流上、中、下游及支流江河、吉尔孟河表土的DNA平均浓度分别为0.65 μg/g、1.48 μg/g、0.51 μg/g、0.42 μg/g和1.03 μg/g。布哈河干流中游和支流吉尔孟河的表土DNA平均浓度明显高于其他河段分组,这可能是地表植被覆盖、河流输移、核酸降解等共同作用的结果。
图2
图2
布哈河流域表土DNA核酸电泳图和浓度分布
注:图b中数字为样品编号。
Fig. 2
The distribution of electrophoretic diagram of nucleic acid and soil DNA concentration in the Buha River catchment
4.2 表土颗粒组成
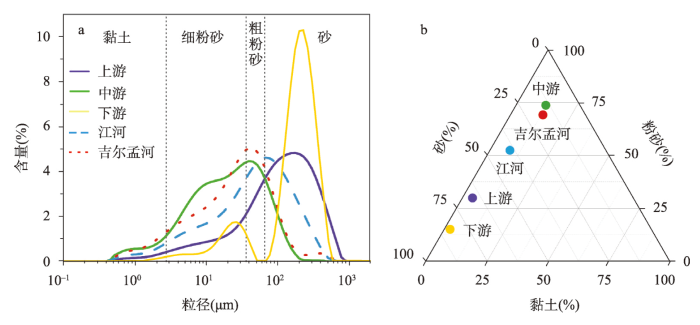
布哈河干流上、中、下游及支流江河、吉尔孟河的粒度特征以各段采样点表土颗粒组成的平均值替代,其结果如图3所示。布哈河流域表土颗粒整体偏粗,平均中值粒径为53.75 μm,以粗粉砂为主。布哈河干流的颗粒组成表现出从上游到下游先变细后变粗特征,这可能是不同物源的结果。具体而言,干流上游河段表土中值粒径为72.9 μm,以砂(> 63 μm)为主,约占50.8 %,黏土含量极低且呈典型单峰状(图3a),属黏土质砂土(图3b);干流中游中值粒径为57.74 μm,以粉砂为主,约占49.4%,峰值出现在粗粉砂粒径范围,也呈单峰状,但黏土和细粉砂含量比上游表土含量高,属砂质黏土(图3b);相较干流上游和中游,下游表土中值粒径为73.95 μm且粉砂和砂各占46%,属砂土(图3b),特别是其粒径分布呈典型的双峰状,在细粉砂和砂范围均出现一个峰值,这表明下游表土可能有两种物质来源。就主要支流而言,江河和吉尔孟河的中值粒径分别为60.65 μm和36.3 μm,表明更靠近下游的吉尔孟河表土比江河的颗粒细,粒级分布均以粉砂为主且呈单峰状(图3a),分别约占58.4 %和48.4 %,均为砂质黏土(图3b)。
图3
图3
布哈河流域表土的颗粒组成
Fig. 3
Particle composition of top soils in the Buha River catchment
4.3 表土元素含量
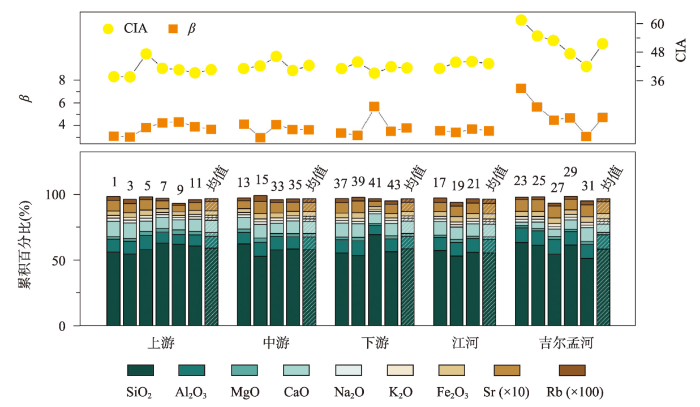
布哈河流域表土的常量元素和微量元素的含量及淋溶系数(β)和化学蚀变指数(CIA)等化学指标的结果如图4所示。总体而言,该区域表土的主要成壤元素如Si、Al、Mg、Ca、Na、K、Fe、Rb、Sr的含量范围分别为:51.48%~69.47%、7%~11.24%、1.37%~3.45%、2.76%~11.8%、1.76%~2.29%、1.45%~2.26%、2.12%~4.22%、0.04‰~0.09‰、0.15‰~0.45‰,平均值分别为58.28%、9.85%、2.25%、8.69%、2.02%、1.99%、3.55%、0.07‰、0.26‰,各元素在布哈河流域内的差异较为明显(图4)。在布哈河干流范围内,表土中Si、Al、Fe、K、Rb等稳定元素含量在上游河段的平均含量分别为59.24%、9.11%、3.28%、1.84%、0.07‰;中游为58.05%、9.71%、3.55%、1.97%、0.07‰;下游为58.81%、9.52%、3.36%、1.92%、0.07‰;支流江河为55.56%、10.32%、3.78%、2.03%、0.08‰;吉尔孟河为58.54%、10.84%、3.91%、2.19%、0.09‰,上述Al、Fe、K、Rb 4类元素含量总体呈现出上游低、中游高、下游偏低的变化趋势,吉尔孟河高于江河,而Si则相反,这与粒度分布较为一致(图3)。易风化的Na、Mg元素含量则与上述五类元素的分布有所不同,在干流上游的含量分别为2.06%、2.63%,干流中游为2.02%、2.57%,干流下游为2.01%、2.1%,支流江河为2%、1.92%,吉尔孟河为1.99%、1.87%,总体表现出由上游到下游逐渐减少的趋势(图4),江河高于吉尔孟河,这可能是不同河段风化程度不一致的结果[52]。而活泼元素Ca、Sr在干流上游的含量分别为9.45%、0.23‰,干流中游为9.17%、0.28‰,干流下游为9.41%、0.26‰,支流江河为9.63%、0.32‰,吉尔孟河为6.24%、0.23‰(图4),Ca元素在干流上的分布趋势总体上与稳定元素相反,而Sr则相同,支流上Ca和Sr的分布基本一致,江河均高于吉尔孟河,这可能是两种元素在表土中的赋存状态、淋溶程度共同作用的结果。进一步地,化学指标β和CIA总体呈相似特征,整个流域中β和CIA最高值均出现在支流吉尔孟河,分别为4.76和51.66,在干流范围内下游β值最高为3.81,中游段的CIA最高为42.45(图4),说明吉尔孟河是布哈河流域风化淋溶程度最高的区域,中游和下游分别是干流风化和淋溶程度最高的区域。
图4
图4
布哈河流域表土的元素含量和化学指标分布
注:图中数字为样品编号。
Fig. 4
The distribution of elements contents and chemical index of top soils in the Buha River catchment
4.4 表土有机质和碳酸盐组成
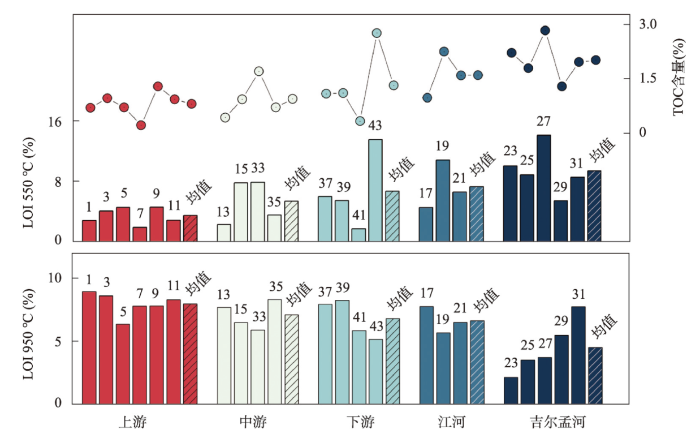
土壤在550 ℃煅烧下的烧失量常被用以指代土壤中的有机质含量,而在950 ℃煅烧下的烧失量则用来代表土壤碳酸盐含量[53]。结果表明,布哈河流域表土550 ℃烧失量反映的有机质含量变化范围为1.79%~14.11%,均值为6.32%;TOC含量在0.23%~2.82%之间,均值为1.31%;碳酸盐含量在2.19%~8.96%之间,均值为6.66%(图5)。有机质含量最高值出现在支流吉尔孟河,碳酸盐含量最高值则出现在布哈河干流上游段。具体而言,有机质和TOC在干流上游的含量分别为3.53%、0.81%,干流中游为5.42%、0.95%,干流下游为6.73%、1.32%,支流江河为7.35%、1.6%,吉尔孟河为9.43%、2.01%,总体表现出由上游到下游逐渐增高的趋势(图5),江河低于吉尔孟河,这可能是不同河段植被覆盖和土地利用不一致的结果,而且这种趋势与Na、Mg等易风化元素的变化趋势相反。而碳酸盐在布哈河干流上游、中游、下游及支流江河和吉尔孟河的含量分别为7.99%、7.13%、6.82%、6.67%、4.55%,也展现出与有机质含量相反的变化趋势(图5)。
图5
图5
布哈河流域表土的烧失量(LOI)和TOC含量分布
注:图中数字为样品编号。
Fig. 5
The distribution of loss on ignition (LOI) and TOC contents of top soils in the Buha River catchment
5 讨论
5.1 布哈河流域表土DNA浓度与理化组成的关系
5.1.1 布哈河流域表土理化特征
基于对布哈河流域干流和主要支流河段表土的系统性采样,结合各土样的粒度、元素、烧失量、TOC等理化指标分析,布哈河流域表土理化组成的空间分布得以体现。就干流而言,① 上游段表土主要由砂和粗粉砂构成,中值粒级达72.9 μm且呈单峰状(图3);赋存于细颗粒物质中的元素Al、Fe、K、Rb、Sr含量较低,富集在粗颗粒中的Si含量最高,而易风化和迁移的Na、Mg、Ca元素含量为流域最高[54],虽因Si含量较高导致淋溶系数β偏高,但化学蚀变指数CIA最低(图4);有机质含量和TOC为整个流域最低,碳酸盐含量最高(图5)。上述理化特征总体反映出该区域的表土颗粒较粗,物质来源单一,有机物来源较少,成壤和风化淋溶作用最弱,总体反映出寒冷干燥的气候特征[44]。② 中游段表土的理化特征:颗粒以粉砂为主,中值粒级为57.74 μm,较上游偏细(图3);Al、Fe、K、Rb、Sr含量为干流最高,Si最低,也说明了该段的细颗粒含量最多,粗颗粒最少,因Si含量显著减少而导致淋溶系数β最低,化学蚀变指数CIA最高(图4);有机质含量和TOC较上游偏高,碳酸盐含量则偏低(图5)。这些特征表明中游区域表土物质来源均一,较上游相对温和湿润、有机物来源增多,成壤和风化淋溶作用增强。③ 下游段表土颗粒最粗,中值粒级为73.95 μm,但在细粉砂区和砂区均出现了峰状(图3),这表明该段表土物源有两种,即细粉砂区的颗粒可能来自于母质的风化和成壤,而砂区的颗粒则可能来自于河流的搬运和堆积;与粒度特征对应的Al、Fe、K、Rb、Sr相较于中游偏低[54-55],而Si则偏高,淋溶系数β最高,化学蚀变指数CIA偏高(图4);有机质含量和TOC为干流段最高,碳酸盐含量则最低(图5)。这些理化特征表明下游区表土物质来源复杂,河道摆动和平坦地势促使了河水中粗颗粒物在下游的堆积,且河流的冲刷和淋溶的加剧使表土中碳酸盐流失,而土壤中的有机质含量在更加温和湿润的环境中得到提升,泥沙堆积也可能带来更多中游段的有机物[39]。
相较于干流段表土的理化特征,位于干流中下游北部的主要支流江河和吉尔孟河的表土理化特征则有所差异。江河的采样点主要集中在该支流的下游段,其粒度、元素及化学指数的组成与干流中游段相似,但有机质和TOC含量明显高于干流中游段,这可能是江河下游段农作物较多和河流搬运堆积的结果。对于更加靠近干流下游的吉尔孟河表土,则颗粒最细,中值粒级仅为36.3 μm,淋溶系数β、化学蚀变指数CIA、有机质和TOC含量为整个布哈河流域最高,而碳酸盐含量最低,这些特征表明吉尔孟河流域与布哈河干流下游段具有不同的地理环境,吉尔孟河在独特的地形下气候更加温和湿润,成壤和风化淋溶作用更强[55]。
5.1.2 表土DNA浓度与理化组成的关系
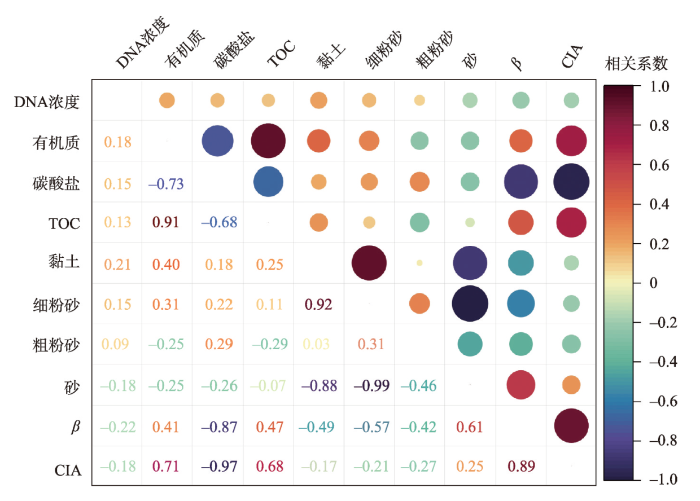
土壤中的DNA通常来源于地表生长的植物残存DNA、活动的动物遗存(如遗体、毛发、粪便和尿液等)DNA和其中活跃的微生物DNA[4],其遗存状况主要受DNA自身特征(如DNA构象、链长等)、非生物环境(如气候、颗粒组成、pH、土地利用方式等)和生物环境(微生物群落等)控制[12,28]。对于土壤自身的理化特征这一非生物环境,前期研究表明DNA浓度与土壤有机质和TOC含量有一定相关性[38],且更易被土壤中的细颗粒物质及黏土矿物吸附[23]。布哈河流域表土DNA与表土有机质、碳酸盐、TOC、颗粒含量、化学指标等理化组成的相关性分析如图6所示。结果表明,在95%置信区间下布哈河表土DNA浓度与有机质含量和TOC的相关性系数分别为0.18和0.13,显示出较弱的正相关性,但也表明表土DNA来源并不完全与有机质一致。相较于DNA这一生物细胞携带的大分子核酸,土壤中的有机物来源和组成更为丰富,动植物残体分解物、微生物分泌物、人为有机肥料及生活垃圾等均可成为土壤中的有机质,且主要由碳水化合物、含氮化合物(蛋白质)、木质素等物质组成[4,38]。生物细胞大分子核酸仅为土壤有机质中比例较低的一部分,且易降解和消失,更多受控于活体生物遗存,因此二者正相关性并不显著。
图6
图6
布哈河流域表土DNA浓度与理化组成的相关性分析
Fig. 6
Correlation analysis between DNA concentration and organic matter, carbonate, TOC, grain size, chemical index of top soil in the Buha River catchment
除DNA浓度与有机质呈一定正相关外,布哈河表土DNA浓度与黏土含量的正相关性最高(r = 0.21),而与砂含量呈负相关性(r = -0.18)(图6)。相关研究表明,黏土等细颗粒物质通过分子间引力和化学键力等物理、化学作用将生物大分子DNA吸附在黏土矿物表面,而粗颗粒物质因低吸附能力和较大孔隙不易截留和吸附生物DNA[23,56]。另一方面,布哈河流域表土DNA浓度与淋溶系数(β)和化学蚀变指数(CIA)均呈反相关,相关系数分别为-0.22和-0.18(图6),表明暖湿环境带来的土壤淋溶风化较显著的降低了表土DNA浓度,可能是生物大分子DNA被降解或迁移的结果。此外,布哈河表土的黏土和粉砂含量总体上与β及CIA指数呈反相关(图6),而砂含量则相反,进一步表明表土成壤过程中伴随着较强的淋溶风化作用[55]。综上,布哈河表土DNA浓度与其理化组成的关系表明,生物大分子DNA通过被细颗粒物质吸附成为土壤有机质的一部分,但也易在淋溶风化作用下被搬运或降解。因此,在选择不同类型地质载体开展沉积物DNA分析之前,需充分考虑流域内沉积物源的成壤环境。需指出的是,布哈河表土DNA浓度与颗粒组成及淋溶风化强度间的相关性虽展现出其指示意义但并不特别显著,可能是采样点较少、未分区域进行空间分析、分析手段(如冗余分析等)不够全面等因素的共同结果。因此,在今后地表过程分析中需进一步改善采样设计和空间分析,进而增加数据分析的多样性和可靠性。
5.2 布哈河流域表土DNA浓度空间分布的环境因素
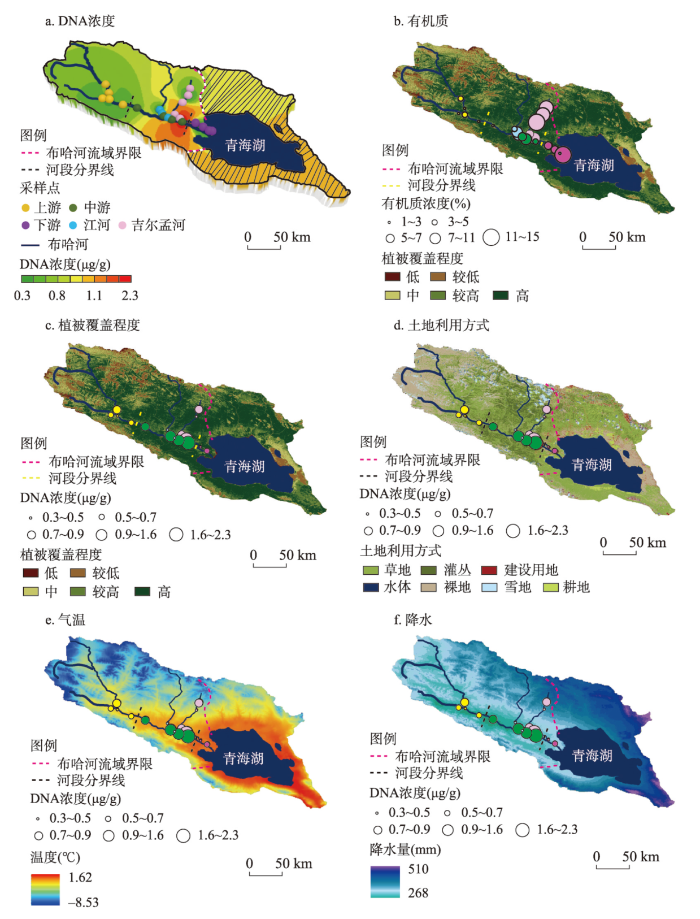
基于布哈河流域干流上、中、下游及主要支流江流、吉尔孟河区域中各采样点的空间分布,利用克里金法将各采样点表土DNA浓度在Arcgis中插值计算,布哈河流域不同河段的表土DNA浓度空间分布如图7a所示。结果表明,布哈河流域表土DNA平均浓度为0.91 μg/g,具体表现为布哈河干流中游段的表土DNA含量最高(1.48 μg/g),支流吉尔孟河次之(1.03 μg/g),干流下游段(0.51 μg/g)和支流江河(0.42 μg/g)最低。另一方面,布哈河流域表土有机质含量的分布表明(图7b),高值主要出现在干流下游段(6.73%)和支流吉尔孟河(9.43%),而干流中游段的有机质含量则较低(5.42%),二者间空间分布的差异也说明有机质含量不能直接反映土壤DNA的浓度变化。
图7
图7
布哈河流域表土DNA浓度、有机质含量、植被覆盖程度、土地利用方式、平均气温和平均降水的空间分布
Fig. 7
Spatial distributions of topsoil DNA concentration, organic matter content, vegetation cover, land use, mean temperature, and mean precipitation in the Buha River catchment
通过分析布哈河流域的植被覆盖程度(图7c)和土地利用方式(图7d),该区域的表土DNA来源及其与土壤有机质分布差异的原因得以体现。相较于植被覆盖率较低的干流上游河段,布哈河干流中下游及江河、吉尔孟河的植被覆盖程度较高,这为土壤中的DNA及有机质提供了丰富的植物来源。另一方面,布哈河流域干流中游段主要以草地和稀疏低矮灌木丛为主,而在支流江河、吉尔孟河则更多以茂密灌木丛为主,上游河段更见裸地。这反映出当地畜牧的主要区域集中在干流中游区域,大量的黄牛、牦牛、羊等畜牧动物频繁活跃在这些区域,而灌木丛内更多见稀疏的野生动物,如高原鼠兔等。这些人类放牧的动物为布哈河干流中下游段提供了大量的动物DNA片段,此外草地内大量细菌、真菌等微生物也贡献了较高的DNA。因此,虽然吉尔孟河表土的有机质含量最高,但其内含的DNA却次于布哈河干流中游段。类似地,地处法国西北部的草地土壤中源自于植物、细菌、真菌中的DNA浓度是植被长久覆盖地区土壤的1.5倍[57]。
相较于植被覆盖程度高且以草地为主的布哈河干流中游段,下游地区除当地居民放牧外,还因季节性农作物耕种而人工施加有机肥,且主要集中在该区域的牧民和游客带来的生活垃圾也增加了土壤有机质含量,因此下游段土壤有机质含量为整个干流区最高。但是,下游区域表土DNA平均浓度仅为0.51 μg/g,表现出与中游段相反的趋势(图7a),这可能是下列几方面因素共同作用的结果。布哈河流域1970—2000年间的年均气温(图7e)和年均降水量(图7f)分布显示干流下游段的温度和降水均高于中游段。较高的温度和较湿润的环境增加了表土中DNA链的降解程度[4],降低了土壤中的DNA浓度。另一方面,地势较平坦的干流下游段在较高降水下易发生河水漫流[58],表土DNA也随降水和漫流现象而被淋溶和流失,进一步降低了该区域表土DNA浓度。此外,干流下游段地表主要是农耕和畜牧交互利用的方式[44]。当地牧民在冬季将家畜圈养在一定区域内喂养;但在夏季随着家畜前往干流中游段散养式放牧,该区域被用以种植青稞、玉米等农作物。Wolinska等[38]指出农耕活动减弱了土壤肥力并通过翻耕等增加了土壤孔隙,促使了土壤中的DNA进一步降解和淋溶。因此,干流下游段的农耕活动也在一定程度上减少了表土DNA浓度[56]。总之,布哈河干流中游河段表土DNA的高含量是较细的土壤质地、高植被覆盖(草地)和放牧活动导致其富集的结果,而下游段则可能是暖湿环境下淋溶、河流输移、农作物耕种3种环境因素共同导致了表土DNA含量的减弱。
6 结论
本文通过对适宜DNA保存的布哈河流域表土DNA浓度和常规理化指标测定,结合气候、植被覆盖、土地利用等环境要素,分析了该流域表土DNA浓度空间分布及影响因素,得出如下结论:
(1)布哈河流域表土DNA平均浓度为0.91 μg/g,其高值主要分布在干流中游及支流吉尔孟河区域,含量分别为1.48 μg/g和1.03 μg/g,而植被覆盖程度高、牧民集中地的下游区域则含量较低,含量仅为0.51 μg/g。
(2)布哈河流域表土DNA浓度与其自身的理化组成有关。黏粒和有机质含量可能是影响土壤DNA浓度的正向因子,而砂含量及风化淋溶作用可能是负向因子,这为开展DNA分析的沉积物载体选择提供了科学依据。
(3)气候、植被覆盖、河流输移、土地利用方式是影响土壤DNA浓度的主要环境因子。植被覆盖程度高、放牧活动频繁有助于DNA在土壤中的富集,而暖湿环境下较强的风化淋溶、河流冲刷、农作物耕种会减弱土壤DNA浓度。
致谢
感谢法国国家科研中心(CNRS)、萨瓦勃朗峰大学EDYTEM实验室Fabien Arnaud研究员、Charline Giguet-Covex研究员、中国科学院南京地理与湖泊研究所林琪特聘研究助理对沉积物DNA实验的指导;感谢圣湘生物科技股份有限公司郭鑫武高级工程师、湖南文理学院生命与环境科学学院林旺副教授对沉积物DNA浓度测定的帮助;感谢青海师范大学田万霞硕士生和西北高原生物研究所李英年研究员对野外采样和常规理化实验分析的支持;特别感谢评审专家和编辑部老师的建设性意见。
参考文献
Ancient DNA indicates human population shifts and admixture in northern and southern China
DOI:10.1126/science.aba0909
PMID:32409524
[本文引用: 1]

Human genetic history in East Asia is poorly understood. To clarify population relationships, we obtained genome-wide data from 26 ancient individuals from northern and southern East Asia spanning 9500 to 300 years ago. Genetic differentiation in this region was higher in the past than the present, which reflects a major episode of admixture involving northern East Asian ancestry spreading across southern East Asia after the Neolithic, thereby transforming the genetic ancestry of southern China. Mainland southern East Asian and Taiwan Strait island samples from the Neolithic show clear connections with modern and ancient individuals with Austronesian-related ancestry, which supports an origin in southern China for proto-Austronesians. Connections among Neolithic coastal groups from Siberia and Japan to Vietnam indicate that migration and gene flow played an important role in the prehistory of coastal Asia.Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Ancient DNA: Achievements in the first 35 years and its prospective
DOI:10.1360/N072015-00539 URL [本文引用: 1]
古DNA研究35年回顾与展望
Evolving ancient DNA techniques and the future of human history
DOI:10.1016/j.cell.2022.06.009
PMID:35868268
[本文引用: 1]

Ancient DNA (aDNA) techniques applied to human genomics have significantly advanced in the past decade, enabling large-scale aDNA research, sometimes independent of human remains. This commentary reviews the major milestones of aDNA techniques and explores future directions to expand the scope of aDNA research and insights into present-day human health.Copyright © 2022 Elsevier Inc. All rights reserved.
Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: Overview and recommendations
The use of lake sedimentary DNA to track the long-term changes in both terrestrial and aquatic biota is a rapidly advancing field in paleoecological research. Although largely applied nowadays, knowledge gaps remain in this field and there is therefore still research to be conducted to ensure the reliability of the sedimentary DNA signal. Building on the most recent literature and seven original case studies, we synthesize the state-of-the-art analytical procedures for effective sampling, extraction, amplification, quantification and/or generation of DNA inventories from sedimentary ancient DNA (sedaDNA) via high-throughput sequencing technologies. We provide recommendations based on current knowledge and best practises.
Sedimentary ancient DNA as a tool in paleoecology
PaleoEcoGen: Unlocking the power of ancient environmental DNA to understand past ecological trends
Holocene vegetation and plant diversity changes in the north-eastern Siberian treeline region from pollen and sedimentary ancient DNA
Collapse of the mammoth-steppe in central Yukon as revealed by ancient environmental DNA
The temporal and spatial coarseness of megafaunal fossil records complicates attempts to to disentangle the relative impacts of climate change, ecosystem restructuring, and human activities associated with the Late Quaternary extinctions. Advances in the extraction and identification of ancient DNA that was shed into the environment and preserved for millennia in sediment now provides a way to augment discontinuous palaeontological assemblages. Here, we present a 30,000-year sedimentary ancient DNA (sedaDNA) record derived from loessal permafrost silts in the Klondike region of Yukon, Canada. We observe a substantial turnover in ecosystem composition between 13,500 and 10,000 calendar years ago with the rise of woody shrubs and the disappearance of the mammoth-steppe (steppe-tundra) ecosystem. We also identify a lingering signal of Equus sp. (North American horse) and Mammuthus primigenius (woolly mammoth) at multiple sites persisting thousands of years after their supposed extinction from the fossil record.© 2021. The Author(s).
Long livestock farming history and human landscape shaping revealed by lake sediment DNA
Pleistocene sediment DNA reveals hominin and faunal turnovers at Denisova Cave
DOI:10.1038/s41586-021-03675-0
[本文引用: 2]

Denisova Cave in southern Siberia is the type locality of the Denisovans, an archaic hominin group who were related to Neanderthals1–4. The dozen hominin remains recovered from the deposits also include Neanderthals5,6 and the child of a Neanderthal and a Denisovan7, which suggests that Denisova Cave was a contact zone between these archaic hominins. However, uncertainties persist about the order in which these groups appeared at the site, the timing and environmental context of hominin occupation, and the association of particular hominin groups with archaeological assemblages5,8–11. Here we report the analysis of DNA from 728 sediment samples that were collected in a grid-like manner from layers dating to the Pleistocene epoch. We retrieved ancient faunal and hominin mitochondrial (mt)DNA from 685 and 175 samples, respectively. The earliest evidence for hominin mtDNA is of Denisovans, and is associated with early Middle Palaeolithic stone tools that were deposited approximately 250,000 to 170,000 years ago; Neanderthal mtDNA first appears towards the end of this period. We detect a turnover in the mtDNA of Denisovans that coincides with changes in the composition of faunal mtDNA, and evidence that Denisovans and Neanderthals occupied the site repeatedly—possibly until, or after, the onset of the Initial Upper Palaeolithic at least 45,000 years ago, when modern human mtDNA is first recorded in the sediments.
Fifty thousand years of Arctic vegetation and megafaunal diet
DOI:10.1038/nature12921 [本文引用: 1]
Ancient plant DNA in lake sediments
DOI:10.1111/nph.14470
PMID:28370025
[本文引用: 2]

Contents 924 I. 925 II. 925 III. 927 IV. 929 V. 930 VI. 930 VII. 931 VIII. 933 IX. 935 X. 936 XI. 938 938 References 938 SUMMARY: Recent advances in sequencing technologies now permit the analyses of plant DNA from fossil samples (ancient plant DNA, plant aDNA), and thus enable the molecular reconstruction of palaeofloras. Hitherto, ancient frozen soils have proved excellent in preserving DNA molecules, and have thus been the most commonly used source of plant aDNA. However, DNA from soil mainly represents taxa growing a few metres from the sampling point. Lakes have larger catchment areas and recent studies have suggested that plant aDNA from lake sediments is a more powerful tool for palaeofloristic reconstruction. Furthermore, lakes can be found globally in nearly all environments, and are therefore not limited to perennially frozen areas. Here, we review the latest approaches and methods for the study of plant aDNA from lake sediments and discuss the progress made up to the present. We argue that aDNA analyses add new and additional perspectives for the study of ancient plant populations and, in time, will provide higher taxonomic resolution and more precise estimation of abundance. Despite this, key questions and challenges remain for such plant aDNA studies. Finally, we provide guidelines on technical issues, including lake selection, and we suggest directions for future research on plant aDNA studies in lake sediments.© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Pollen, macrofossils and sedaDNA reveal climate and land use impacts on Holocene mountain vegetation of the Lepontine Alps, Italy
Homogenization of lake cyanobacterial communities over a century of climate change and eutrophication
Sedimentary ancient DNA reveals a threat of warming-induced alpine habitat loss to Tibetan Plateau plant diversity
Studies along elevational gradients worldwide usually find the highest plant taxa richness in mid-elevation forest belts. Hence, an increase in upper elevation diversity is expected in the course of warming-related treeline rise. Here, we use a time-series approach to infer past taxa richness from sedimentary ancient DNA from the south-eastern Tibetan Plateau over the last ~18,000 years. We find the highest total plant taxa richness during the cool phase after glacier retreat when the area contained extensive and diverse alpine habitats (14-10 ka); followed by a decline when forests expanded during the warm early- to mid-Holocene (10-3.6 ka). Livestock grazing since 3.6 ka promoted plant taxa richness only weakly. Based on these inferred dependencies, our simulation yields a substantive decrease in plant taxa richness in response to warming-related alpine habitat loss over the next centuries. Accordingly, efforts of Tibetan biodiversity conservation should include conclusions from palaeoecological evidence.
Long-term fungus-plant covariation from multi-site sedimentary ancient DNA metabarcoding
DNA from lake sediments reveals long-term ecosystem changes after a biological invasion
Late Quaternary dynamics of Arctic biota from ancient environmental genomics
DOI:10.1038/s41586-021-04016-x
[本文引用: 1]

During the last glacial–interglacial cycle, Arctic biotas experienced substantial climatic changes, yet the nature, extent and rate of their responses are not fully understood1–8. Here we report a large-scale environmental DNA metagenomic study of ancient plant and mammal communities, analysing 535 permafrost and lake sediment samples from across the Arctic spanning the past 50,000 years. Furthermore, we present 1,541 contemporary plant genome assemblies that were generated as reference sequences. Our study provides several insights into the long-term dynamics of the Arctic biota at the circumpolar and regional scales. Our key findings include: (1) a relatively homogeneous steppe–tundra flora dominated the Arctic during the Last Glacial Maximum, followed by regional divergence of vegetation during the Holocene epoch; (2) certain grazing animals consistently co-occurred in space and time; (3) humans appear to have been a minor factor in driving animal distributions; (4) higher effective precipitation, as well as an increase in the proportion of wetland plants, show negative effects on animal diversity; (5) the persistence of the steppe–tundra vegetation in northern Siberia enabled the late survival of several now-extinct megafauna species, including the woolly mammoth until 3.9 ± 0.2 thousand years ago (ka) and the woolly rhinoceros until 9.8 ± 0.2 ka; and (6) phylogenetic analysis of mammoth environmental DNA reveals a previously unsampled mitochondrial lineage. Our findings highlight the power of ancient environmental metagenomics analyses to advance understanding of population histories and long-term ecological dynamics.
Pastoralism increased vulnerability of a subalpine catchment to flood hazard through changing soil properties
Ancient plant DNA in archaeobotany
DOI:10.1007/s00334-007-0125-7 URL [本文引用: 1]
Sedimentary DNA and molecular evidence for early human occupation of the Faroe Islands
Ancient environmental DNA and environmental archaeology
古环境DNA与环境考古
A 2-million-year-old ecosystem in Greenland uncovered by environmental DNA
Applications of ancient DNA preserved in sediment in paleo-ecology, paleo-environment and paleo-climate studies
沉积物中古DNA在古生态、古环境和古气候研究中的应用
DOI:10.13745/j.esf.2017.02.028
[本文引用: 1]

随着分子生物学技术的发展,DNA技术的应用逐渐从生物学研究拓展到进化、环境和气候变化等研究领域。因为DNA容易受到水解、氧化、紫外辐射、酶解等过程的破坏,所以人们认为DNA很容易降解,保存时间不长,但是近年来的研究成果显示,古生物和古人类的化石或骨骼、冻土、湖泊海洋沉积物等样品中包含已保存数千年甚至长达数百万年以来的古DNA。古DNA作为重要的信息载体,可以诠释人类、动植物和微生物进化和迁移过程及它们对环境和气候变化的响应。反过来,古DNA在古环境、古气候重建方面也起着重要作用,沉积物样品的古DNA往往保存了历史时期的水体和水体周围环境以及气候变化等信息。通过对沉积物中古DNA的研究,可以获得历史时期的水生生物(包括藻类、水生动植物等)的群落演化,和水体周围岸基生活的动植物群落(植物落叶、动物毛发冲带到水体并沉积下来)演替,进而反演水体和水体周围气候环境以及气候变化信息。通过研究古人类和动物化石中保存的DNA,可以探讨人类的迁徙和进化过程,溯源家畜的驯化历史。本文将介绍环境古DNA的保存和可靠性评价,重点探讨古DNA在古生态以及古环境、古气候重建研究中的优势及应用,并展望古DNA在生物进化和环境研究等领域的应用前景。
Application of modern molecular biotechnology in studying ancient DNA of lake sediments
In recent years, the development of new techniques in molecular biology offers a new way for understanding the evolution of lake ecosystem. Ancient DNA could well record the migration and evolution of animals, plants, algae and even microbes, and thus has become an effective tool to reconstruct paleoclimate and paleoenvironment. Studies on the ancient DNA preserved in lake sediments have provided abundant information and high-resolution data that are needed for reconstruction of ancient microbial, plant, or animal community, as well as the ancient environmental conditions. Here, we firstly introduced the preservation characteristics of the ancient DNA and DNA extraction methods from lake sediments, and then reviewed the latest molecular biology techniques for studying ancient DNA preserved in lake sediments. We also discussed the application of ancient DNA in lake sediments including reconstruction of paleoclimate, paleoecology and the impacts of human activities on paleoenvironment. Finally, some problems and possible trends on the research of ancient DNA in lake sediments were summarized.
现代分子生物技术在湖泊沉积物古DNA研究中的应用
分子生物学技术的发展,为从分子生物学角度探讨湖泊生态系统演化提供了一个新窗口。依据湖泊沉积物中古DNA研究,从古微生物、动植物群落结构组成及数量变化等方面开展研究,可进一步反演湖泊及周边的气候环境变化,探讨生物与气候、人类活动之间的关系。本文首先介绍了湖泊沉积物中古DNA来源、保存特点以及提取方法,其次比较分析了现代分子生物技术在湖泊沉积物古DNA中的应用特点,并阐述了湖泊沉积物古DNA在重建古气候、古生态以及人类活动对古环境影响方面的应用,最后对目前湖泊沉积物古DNA研究方法存在的一些问题进行了总结。
Progress in the application of lake sediment DNA in climate and environmental change and ecosystem response
DOI:10.18307/2021.0303 URL [本文引用: 1]
湖泊沉积物DNA在气候环境变化和生态系统响应研究中的应用
Modern processes of lacustrine plant sedimentary ancient DNA
DOI:10.17521/cjpe.2021.0386
[本文引用: 1]

Study on paleovegetation is with great significance on understanding the modern vegetation composition, distribution, and its responses to global change. During recent decades, it has been a rapidly developing field in molecular palaeoecology to track the long-term vegetation composition and plant diversity changes by using sedimentary ancient DNA. Meanwhile, the lacustrine sedimentary ancient plant DNA (PsedaDNA) is becoming a popular proxy for palaeovegetation and palaeoecological studies. However, the modern process of PsedaDNA remains unclear compared with pollen analysis, which limits its further application. Here, we reviewed the PsedaDNA researches and tried to elucidate the modern processes of PsedaDNA, including the source, deposition, and preservation processes, and to understand the relationships between the PsedaDNA and modern vegetation. PsedaDNA is mainly derived from the lake surroundings or within the catchment. The abundance and composition of PsedaDNA are influenced by the biomass of terrestrial plants, and factors such as DNA degradation, the adsorption, and dilution of particles in soils and sediments during the transportation and deposition. Moreover, the preservation of PsedaDNA is largely affected by both biotic and abiotic factors, including the microbial activity, chemical properties of lake water (conductivity and pH), and sediment composition. PsedaDNA can be used to reveal the contemporary vegetation and climate information but was incompetent to quantitatively palaeovegetation reconstruction. Considering the complexity of modern processes, the interpretation of PsedaDNA results should be done with extra caution. To sum up, PsedaDNA is still in its infancy, which has been benefiting from the progress of molecular biotechnology, optimization of experimental design, species barcodes, and sophistication of reference databases. The PsedaDNA technique including DNA metabarcoding and metagenomics will certainly promote the development of Chinese palaeophytoecology research.
湖泊沉积植物古DNA的现代过程
DOI:10.17521/cjpe.2021.0386
[本文引用: 1]

对植被历史变化过程的研究是理解现代植被组成、分布及其对全球变化响应的基础。近年来, 随着分子古生态学的发展, 分析沉积介质中的陆生植物古DNA信号, 以研究植被及植物多样性演变的历史过程正在成为研究热点, 湖泊沉积植物古DNA已成为古植被和古生态学研究的成熟代用指标。然而与第四纪孢粉分析相比较, 湖泊沉积植物古DNA的现代过程依然不明确, 成为其进一步发展和应用的限制因素。基于此, 该文综述了湖泊沉积植物古DNA技术研究进展, 尝试阐明湖泊沉积植物古DNA的现代过程, 包括植物DNA的来源、沉积和保存过程及其影响因素, 以及植物DNA与现代植被的关系等。已有研究表明, 湖泊沉积植物古DNA主要来自湖泊周边或流域范围, 其丰度和组成除受到源植物生物量的影响外, 同样受到沉积物的搬运和沉积过程中DNA降解作用、土壤以及沉积物中颗粒的吸附过程和稀释作用等因素的影响。湖泊沉积物中植物DNA的保存则主要受到微生物活动、湖水的化学性质(电导率和pH值)、湖泊深度、沉积物组成等一系列生物与非生物因素的共同影响。湖泊沉积植物古DNA可以揭示其沉积时代的植物群落类型以及气候环境信息, 但目前并不能够用来定量重建古植被变化过程。鉴于湖泊沉积植物古DNA现代过程的复杂性, 对研究结果的解释要格外小心。与孢粉分析相比, 湖泊沉积植物古DNA研究仍处于起步阶段, 但随着分子生物技术的进步、实验设计的优化、物种条形码的扩充及参考数据库的完善等, 以DNA宏条形码和宏基因组学为主要技术手段的植物古DNA技术, 必将推动我国植物古生态研究的进一步发展。
Lake sedimentary ancient DNA (sedaDNA) reveals paleoclimatic change and ecological evolution
湖泊沉积物古DNA揭示气候环境变化和生态演化
Reconstruction of flora and fauna community evolution and human-earth interaction using sedimentary ancient DNA: Progress and perspectives
沉积物古DNA重构动植物群落变化及人地关系的研究进展
Plant sedimentary DNA as a proxy for vegetation reconstruction in eastern and northern Asia
Vegetation reconstruction from Siberia and the Tibetan Plateau using modern analogue technique: Comparing sedimentary (ancient) DNA and pollen data
To reconstruct past vegetation from pollen or, more recently, lake sedimentary DNA (sedDNA) data is a common goal in palaeoecology. To overcome the bias of a researcher’s subjective assessment and to assign past assemblages to modern vegetation types quantitatively, the modern analogue technique (MAT) is often used for vegetation reconstruction. However, a rigorous comparison of MAT-derived pollen-based and sedDNA-based vegetation reconstruction is lacking. Here, we assess the dissimilarity between modern taxa assemblages from lake surface-sediments and fossil taxa assemblages from four lake sediment cores from the south-eastern Tibetan Plateau and northern Siberia using receiver operating characteristic (ROC) curves, ordination methods, and Procrustes analyses. Modern sedDNA samples from 190 lakes and pollen samples from 136 lakes were collected from a variety of vegetation types. Our results show that more modern analogues are found with sedDNA than pollen when applying similarly derived thresholds. In particular, there are few modern pollen analogues for open vegetation such as alpine or arctic tundra, limiting the ability of treeline shifts to be clearly reconstructed. In contrast, the shifts in the main vegetation communities are well captured by sedimentary ancient DNA (sedaDNA). For example, pronounced shifts from late-glacial alpine meadow/steppe to early–mid-Holocene coniferous forests to late Holocene Tibetan shrubland vegetation types are reconstructed for Lake Naleng on the south-eastern Tibetan Plateau. Procrustes and PROTEST analyses reveal that intertaxa relationships inferred from modern sedaDNA datasets align with past relationships generally, while intertaxa relationships derived from modern pollen spectra are mostly significantly different from fossil pollen relationships. Overall, we conclude that a quantitative sedaDNA-based vegetation reconstruction using MAT is more reliable than a pollen-based reconstruction, probably because of the more straightforward taphonomy that can relate sedDNA assemblages to the vegetation surrounding the lake.
Preservation of sedimentary plant DNA is related to lake water chemistry
DOI:10.1002/edn3.259
URL
[本文引用: 3]

Little is currently known about preservation of plant DNA in lake sediments. Most prior information originates from laboratory experiments while systematic field‐based studies are still lacking. Here, we used the “g” and “h” universal primers for the P6 loop region of the chloroplast trnL (UAA) intron to amplify plant DNA from 219 lake surface sediments from China and Siberia. We introduce (i) the percentage of sequence counts with the best identity ≥95%, (ii) weighted average identity, (iii) weighted average DNA fragment length, and iv) rarefied richness of terrestrial seed plants of plant DNA metabarcoding as proxies for sedimentary DNA preservation and relate them to five environmental variables (lake water conductivity, lake water pH, mean July air temperature, and sampling depth, lake size) using boosted regression tree (BRT) analyses. Our results suggest that lake water chemical characteristics, that is, electrical conductivity and pH, are the most important variables for the preservation of plant DNA in lake sediments. Intermediate water conductivities (100–500 μS cm−1) and neutral to slightly alkaline water pH (7–9) may facilitate plant DNA preservation. Furthermore, deep lakes seem to support plant DNA preservation as indicated by relatively high rarefied richness. We also find high rarefied richness in small lakes compared with large lakes, but this result needs to be assessed by more studies in the future. None of our BRT models shows that mean July air temperature is a key variable to limit plant DNA preservation. To conclude, our results suggest that sedimentary DNA studies can preferentially select deep lakes characterized by intermediate water conductivities and neutral to slightly alkaline pH conditions.
Indica rice spread to the Tibetan Plateau in the 700s CE
DOI:10.1360/SSTe-2021-0349 URL [本文引用: 1]
公元8世纪前后籼稻向青藏高原南部的传播
Food resources of the Khog Gzung site on the Tibetan Plateau revealed by sedimentary ancient DNA
DOI:10.1360/SSTe-2022-0225 URL [本文引用: 1]
堆积物古DNA揭示西藏廓雄遗址的食物构成
Ancient DNA reveals potentially toxic cyanobacteria increasing with climate change
New insights on lake sediment DNA from the catchment: Importance of taphonomic and analytical issues on the record quality
Over the last decade, an increasing number of studies have used lake sediment DNA to trace past landscape changes, agricultural activities or human presence. However, the processes responsible for lake sediment formation and sediment properties might affect DNA records via taphonomic and analytical processes. It is crucial to understand these processes to ensure reliable interpretations for "palaeo" studies. Here, we combined plant and mammal DNA metabarcoding analyses with sedimentological and geochemical analyses from three lake-catchment systems that are characterised by different erosion dynamics. The new insights derived from this approach elucidate and assess issues relating to DNA sources and transfer processes. The sources of eroded materials strongly affect the "catchment-DNA" concentration in the sediments. For instance, erosion of upper organic and organo-mineral soil horizons provides a higher amount of plant DNA in lake sediments than deep horizons, bare soils or glacial flours. Moreover, high erosion rates, along with a well-developed hydrographic network, are proposed as factors positively affecting the representation of the catchment flora. The development of open and agricultural landscapes, which favour the erosion, could thus bias the reconstructed landscape trajectory but help the record of these human activities. Regarding domestic animals, pastoral practices and animal behaviour might affect their DNA record because they control the type of source of DNA ("point" vs. "diffuse").
How have studies of ancient DNA from sediments contributed to the reconstruction of Quaternary floras
DOI:10.1111/nph.13657
PMID:26402315
[本文引用: 1]

499 I. 499 II. 500 III. 500 IV. 500 V. 500 VI. 501 VII. 502 VIII. 504 504 References 505 SUMMARY: Ancient DNA (aDNA) from lake sediments, peats, permafrost soils, preserved megafaunal gut contents and coprolites has been used to reconstruct late-Quaternary floras. aDNA is either used alone for floristic reconstruction or compared with pollen and/or macrofossil results. In comparative studies, aDNA may complement pollen and macrofossil analyses by increasing the number of taxa found. We discuss the relative contributions of each fossil group to taxon richness and the number of unique taxa found, and situations in which aDNA has refined pollen identifications. Pressing problems in aDNA studies are contamination and ignorance about taphonomy (transportation, incorporation, and preservation in sediments). Progress requires that these problems are reduced to allow aDNA to reach its full potential contribution to reconstructions of Quaternary floras. © 2015 The Authors. New Phytologist © 2015 New Phytologist Trust.
The effect of environmental factors on total soil DNA content and dehydrogenase activity
DOI:10.2298/ABS140120013W
URL
[本文引用: 4]

The aim of the study was a statistical evaluation of the impact of selected\n soil factors ? water potential (pF), total organic carbon content (TOC) and\n land use ? on the total DNA content and dehydrogenase activity (DHA) in\n Mollic Gleysol. Additionally, we wanted to establish the interrelations\n between two of the most important biological parameters in soli: activity of\n intracellular dehydrogenases and total DNA content. Soil originating from the\n surface layer of the control site displayed higher DHA (c.a. by 57%) and DNA\n content (c.a. by 25%) as compared to an cultiavted meadow. Our results also\n indicate a significant (p <0.05) positive relationship between the soil DNA\n content and DHA. Importantly, intensive and systematical agricultural soil\n usage resulted in the reduction of its DHA and DNA content.
Controls on seasonal erosion behavior and potential increase in sediment evacuation in the warming Tibetan Plateau
Hotspots of riverine greenhouse gas (CH4, CO2, N2O) emissions from Qinghai Lake Basin on the northeast Tibetan Plateau
Mineralogy of the otoliths of naked carp Gymnocypris przewalskii (Kessler) from Lake Qinghai and its Sr/Ca potential implications for migratory pattern
DOI:10.1360/zd-2012-42-8-1210 URL [本文引用: 1]
青海湖裸鲤(湟鱼)耳石的矿物组成及其Sr/Ca对洄游习性的潜在示踪
Effects of molybdenosis on antioxidant capacity in endangered Przewalski's Gazelles in the Qinghai Lake National Nature Reserve in the northwestern China
Evaluation of changes in ecological security in China's Qinghai Lake Basin from 2000 to 2013 and the relationship to land use and climate change
DOI:10.1007/s12665-013-2955-1 URL [本文引用: 1]
Assessing the impact of human activities on surface pollen assemblages in Qinghai Lake Basin, China
DOI:10.1002/jqs.v33.6 URL [本文引用: 3]
Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies
DOI:10.1111/j.1365-294X.2011.05317.x
PMID:22300434
[本文引用: 1]

DNA metabarcoding refers to the DNA-based identification of multiple species from a single complex and degraded environmental sample. We developed new sampling and extraction protocols suitable for DNA metabarcoding analyses targeting soil extracellular DNA. The proposed sampling protocol has been designed to reduce, as much as possible, the influence of local heterogeneity by processing a large amount of soil resulting from the mixing of many different cores. The DNA extraction is based on the use of saturated phosphate buffer. The sampling and extraction protocols were validated first by analysing plant DNA from a set of 12 plots corresponding to four plant communities in alpine meadows, and, second, by conducting pilot experiments on fungi and earthworms. The results of the validation experiments clearly demonstrated that sound biological information can be retrieved when following these sampling and extraction procedures. Such a protocol can be implemented at any time of the year without any preliminary knowledge of specific types of organisms during the sampling. It offers the opportunity to analyse all groups of organisms using a single sampling/extraction procedure and opens the possibility to fully standardize biodiversity surveys.© 2012 Blackwell Publishing Ltd.
Extracellular DNA extraction
Extracellular DNA extraction from sediment using phosphate buffer and NucleoSpin® Soil kit (MACHEREY NAGEL)
The factors affecting the reproducibility of micro-volume DNA mass quantification in Nanodrop 2000 spectrophotometer
DOI:10.1016/j.ijleo.2017.08.031 URL [本文引用: 1]
Characteristics of surface sediments on the climbing dunes in Shannan wide valley section of Yarlung Tsangpo River, China
DOI:10.7522/j.issn.1000-694X.2021.00176
[本文引用: 1]

In order to reveal the formation process, material source and sedimentological significance of the climbing dunes in the Shannan wide valley section of Yarlung Tsangpo River, cross-sectional sampling was conducted on the climbing dunes of Langsailing, and the particle size and geochemical elemental characteristics of the climbing dunes sediments in different topography parts were analyzed. The results show that the grain size composition of the surface sediments of the climibing dunes is mainly fine and medium sands, while the river floodplain and terraces show obvious wind-water interaction, and the windward and leeward slopes show obvious aeolian characteristics; The particle size characteristics are different in different terrain. The particle size gradually becomes finer and better sorted from the riverbed to upwind slope, while the opposite trend is observed from upwind slope to the leeward slope. The geochemical element composition of the sediments is dominated by Si, Al, Fe and Na, and the small difference in the climbing dune section indicates that they have similar depositional environment. Compared with the upper continental crust (UCC), all the elements are deficient except Si. The A-CN-K ternary diagram and CIA index reveal that chemical weathering of the surface sediments is in the early stage, while the A-CNK-FM diagram indicates that the spatial distributions of Fe and Mg elements are spatially different, which is the result of sorting action of wind. From the chemical weathering index perspective, the weathering of the sediment is enhanced from the river bed to the windward slope. In summary, material source and wind dynamics are important factors leading to the spatial differences in grain size and chemical elements. Therefore, we determine the climbing dunes of Langsailing are near-source dunes, and the material source is mainly from the river sediments of the Yarlung Tsangpo River. Meanwhile, the variance of sediment characteristics at different topography part are the active response to the interaction of wind and water.
雅鲁藏布江山南宽谷段爬坡沙丘表层沉积物特征
DOI:10.7522/j.issn.1000-694X.2021.00176
[本文引用: 1]

为揭示雅鲁藏布江山南宽谷段爬坡沙丘的形成过程、物源及沉积学意义,对山南宽谷段朗赛岭爬坡沙丘进行断面采样,分析爬坡沙丘不同地貌部位沉积物的粒度和地球化学元素特征。结果表明:爬坡沙丘表层沉积物以细沙和中沙为主,河漫滩和阶地表现出明显的风水交互特点,迎风坡和背风坡则表现为明显的风成特点;不同地貌部位沉积物粒度特征不同,从河漫滩到迎风坡坡顶,沉积物粒度整体变细,分选逐渐变好,从迎风坡顶到背风坡,分选变差。沉积物的地球化学元素以Si、Al、Fe、Na为主,在不同地貌部位分布差异较小,表明其具有相似的沉积环境,和上陆壳(UCC)相比,除Si以外其余常量元素均有亏损。微量元素集中分布于河漫滩沉积物中,部分存在于稳定矿物中的微量元素可分布于坡麓地带。A-CN-K图解及CIA值揭示了朗赛岭爬坡沙丘处于化学风化的初期,A-CNK-FM图则表明Fe、Mg元素在空间上存在差异,是差异风力风选的结果。从化学风化指标来看,从河漫滩到迎风坡,沉积物的风化程度增强。综上所述,物源和风动力是导致爬坡沙丘粒度和化学元素存在空间差异的重要因素,朗赛岭爬坡沙丘属于近源沙丘,沙源主要是雅鲁藏布江河流相沉积物,不同地貌部位沉积物特征的差异性是对风、水交互作用的积极响应。
Capability of loss-on-ignition as a predictor of total organic carbon in non-calcareous Forest Soils
DOI:10.1080/00103620500306080 URL [本文引用: 1]
Seasonally chemical weathering and CO2 consumption flux of Lake Qinghai river system in the northeastern Tibetan Plateau
DOI:10.1007/s12665-009-0027-3 URL [本文引用: 1]
Variation in loss on ignition of the Nilka loess section in the Yili basin and its impact factors
伊犁尼勒克剖面烧失量变化特征及影响因素
Features and geochemical identification index of deposition couplets in landslide-dammed reservoirs on Loess Plateau of China
黄土高原聚湫沉积旋回特征及地球化学划分
Elemental compositions of the late Quaternary loess-paleosol on the northeastern Qinghai-Tibet Plateau and their implications for provenance
DOI:10.7522/j.issn.1000-694X.2020.00078
[本文引用: 3]

Late Quaternary loess-paleosol deposits on the northeastern Qinghai-Tibet Plateau (NE QTP) recorded information of environmental changes in this area. Previous studies have been carried out on their source area. However, due to the insufficient space coverage of samples in previous studies, this issue still needs further study. We collected samples of loess-paleosol, eolian sands, fluvial sediments and lacustrine deposits on the NE QTP. Chemical compositions of these samples (less than 75 μm fraction) were determined by X-ray fluorescence (XRF) spectrometry. The results show that: (1) The chemical composition of the late Quaternary loess-paleosol on the NE QTP is mainly composed of SiO2, Al2O3, Fe2O3, and CaO; compared with the composition of UCC (upper continental crust), the late Quaternary loess-paleosol on the NE QTP has higher CaO and MgO content, lower Na2O and K2O content. (2) K2O/Al2O3 (molar ratio), TiO2/Al2O3 (molar ratio), Zr/Nb and Zr/Ti ratios show that the source area of the late Quaternary loess-paleosol in the Qinghai Lake area on the NE QTP is different from that of the fluvial sediments and lacustrine sediments in this area, indicating that the loess-paleosol in the Qinghai Lake area is probably from a region far away from the Qinghai Lake area. (3) K2O/Al2O3 (molar ratio), TiO2/Al2O3 (molar ratio), Zr/Nb and Zr/Ti ratios show that the late Quaternary loess-paleosol on the NE QTP and the eolian sand, fluvial sediments and lacustrine deposits in the Qaidam Basin cannot be distinguished, indicating that the weathered detrital material in the Qaidam Basin is likely to provide substantial material for loess-paleosol on the NE QTP under the influence of the near-surface northwest wind and the Westerly circulation.
青藏高原东北部晚第四纪黄土—古土壤的元素组成及其物源指示
DOI:10.7522/j.issn.1000-694X.2020.00078
[本文引用: 3]

青藏高原东北部的晚第四纪黄土-古土壤记录了该区环境变化的信息,以往的物源研究样品空间覆盖度不足,仍需进一步研究。在青藏高原东北部较大范围地采集了黄土-古土壤、风成砂、河流沉积、湖相沉积样品,并对这些样品小于75 μm组分的元素组成进行了X射线荧光光谱(XRF)分析。结果表明:(1)青藏高原东北部晚第四纪黄土-古土壤的元素组成以SiO<sub>2</sub>、Al<sub>2</sub>O<sub>3</sub>、Fe<sub>2</sub>O<sub>3</sub>、CaO为主;与UCC(上地壳)相比,青藏高原东北部晚第四纪黄土-古土壤的CaO、MgO含量较高,Na<sub>2</sub>O、K<sub>2</sub>O含量较低。(2)具有物源指示意义的K<sub>2</sub>O/Al<sub>2</sub>O<sub>3</sub>(摩尔比)、TiO<sub>2</sub>/Al<sub>2</sub>O<sub>3</sub>(摩尔比)、Zr/Nb和Zr/Ti比值图解显示青藏高原东北部青海湖地区的晚第四纪黄土-古土壤与当地的河流沉积、湖相沉积存在较大差异,揭示它们来自青海湖地区以外的区域。(3)K<sub>2</sub>O/Al<sub>2</sub>O<sub>3</sub>(摩尔比)、TiO<sub>2</sub>/Al<sub>2</sub>O<sub>3</sub>(摩尔比)、Zr/Nb和Zr/Ti比值图解显示青藏高原东北部的晚第四纪黄土-古土壤与柴达木盆地的风成砂、河流沉积和湖相沉积重叠在一起,表明柴达木盆地的风化细碎屑物质在近地面西北风、高空西风环流的作用下很可能为青藏高原东北部黄土-古土壤的堆积提供了主要物源。
Persistence of environmental DNA in cultivated soils: Implication of this memory effect for reconstructing the dynamics of land use and cover changes
eDNA refers to DNA extracted from an environmental sample with the goal of identifying the occurrence of past or current biological communities in aquatic and terrestrial environments. However, there is currently a lack of knowledge regarding the soil memory effect and its potential impact on lake sediment eDNA records. To investigate this issue, two contrasted sites located in cultivated environments in France were studied. In the first site, soil samples were collected (n = 30) in plots for which the crop rotation history was documented since 1975. In the second site, samples were collected (n = 40) to compare the abundance of currently observed taxa versus detected taxa in cropland and other land uses. The results showed that the last cultivated crop was detected in 100% of the samples as the most abundant. In addition, weeds were the most abundant taxa identified in both sites. Overall, these results illustrate the potential of eDNA analyses for identifying the recent (< 10 years) land cover history of soils and outline the detection of different taxa in cultivated plots. The capacity of detection of plant species grown on soils delivering sediments to lacustrine systems is promising to improve our understanding of sediment transfer processes over short timescales.
Fungal, bacterial and plant dsDNA contributions to soil total DNA extracted from silty soils under different farming practices: Relationships with chloroform-labile carbon
DOI:10.1016/j.soilbio.2010.11.012 URL [本文引用: 1]
Isotope and hydrochemistry reveal evolutionary processes of lake water in Qinghai Lake
DOI:10.1016/j.jglr.2016.02.007 URL [本文引用: 2]